Abstract
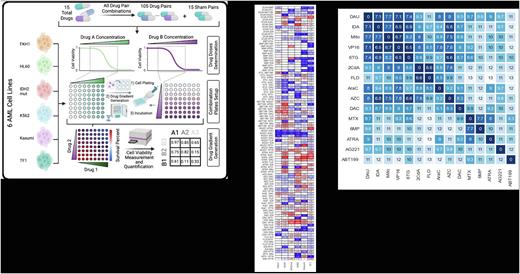
Since the beginning of chemotherapy exist in medical world, few substantial therapeutic changes have been made for patients with acute myeloid leukemia (AML). In the last decade, remarkable improvements have been seen in treatment of AML with several new drugs receiving regulatory approval. Despite promising number of drugs and studies, the outcomes of patients with AML remain unsatisfactory. Even though these drugs passed the trial phases before clinical usage, there are still unexplained areas how drugs interact with each other on each patient who has different genetic profile and biological variations. Therefore, this study was designed for understanding how each drug works on each cell line, drug binary combinations show which synergy model (synergistic, antagonistic or additive), and if the models show a variation with same drug combination on different cell lines, how much strong is their combination effect. With this design, thirteen drugs and two targeted therapies were used in treatments of 5 cell lines. The results of single agent treatments of each cell line were used to determine the drug dosages and parameters of eight-by-eight binary combinations and eleven-by-eleven binary combinations with each drug.
Daunorubicin, cytarabine, idarubicin, mitoxantrone, etoposide as elements of traditional AML treatment were consistent with their clinical reputation as they are part of the first line treatment. The rest of the drugs showed diverse range of response to treatment. Some of the drugs' single dose responses showed incomplete killing effect on AML cell lines but they still have impressive combination effect with some specific drugs. Some unusual treatment elements showed significant effect with specific combination on specific cell lines. An accepted treatment combination which is well known combination in clinic showed reverse effect on a cell line. A couple drug which are already in clinical usage before stem cell transplantation has an adverse interaction with its pair on a cell line.
Both Loewe's Combination Index and Bliss over-excess synergy scores exhibited significant variability with regards to cell line. Clustering of drugs based on their synergy scores across all cell lines and drug combinations resulted in groups that segregated based on drug mechanism, yielding validity to our results. Additionally, we observed that some drugs behave very differently with regards to their synergistic potential from the rest of drugs in their class. A few drugs showed different type of behavior on different cell lines. Some combinations reflected strong synergism or antagonism. In conclusion, the results revealed multiple new potential synergistic drug pairs that could be very beneficial in the clinic as well as illuminate the necessity to account for the disease's heterogeneity not only when considering individual therapeutics but combination of them as well. And a few drugs showed strong antagonism on some combinations which are observed on only specific cell lines. Both synergistic and antagonistic responses are substantial candidates for new animal models and then clinical trials.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal